
Analysis
10 plates per device
Lower variance & higher throughput
Resilient to biological noise
The purpose of this study was to compare the turbidity measurements done by the Reshape Imaging Device (RID) to conventional OD600 measurements done using a plate reader.
The RID allows for higher bulk throughout, although with a lower resolution than that seen for the plate reader. Additionally, the visual inspection of the wells in the RID, allows for further characterization than that of the plate reader.
To summarize, each method for measuring has both pros and cons, the primary ones to be found concern sensitivity and throughput (See table under point 8).
Other applications
Fluorescence
Automate fluorescence assays in any plate format with Reshape and increase throughput
.gif)
Seed germination
Automatically analyze complex phenotypes like germination rate, root growth, and time-to-germination with Reshape
.gif)
.gif)
Introduction
Establishing a laboratory in the early stages of a biotech startup is often met with significant financial and logistical barriers. High costs associated with specialized analytical instruments—particularly those used for essential assays like cell counting—can restrict access to data-driven experimentation, hindering rapid iteration and innovation. Among such instruments, cell quantification systems based on optical density (OD) at 600 nm are widely considered the gold standard due to their reliability and established place in microbiological protocols. However, these systems typically require costly, dedicated spectrophotometers, presenting a steep entry point for resource-constrained startups.
Turbidity-based imaging systems have emerged as a more accessible alternative, often available as part of pay-per-use platforms or lower-cost hardware solutions. Validating these systems against OD600-based methods could democratize early-stage experimental workflows, enabling broader access to reliable biological measurements without prohibitive upfront investments.
In this study, we compare two such systems—one based on turbidity imaging and the other usingOD600—to assess their accuracy and practicality in quantifying cell densities. By examining their performance side-by-side, we aim to evaluate whether lower-cost or usage-based alternatives can offer sufficient reliability to support early-stage research. This validation is not just a technical comparison—it represents a step toward reducing barriers to innovation, empowering emerging biotech companies, and ultimately, accelerating the development of technologies that could benefit biotechnology on a global scale.
Materials and methods
A single colony was inoculated into 1 mL of brain heart infusion agar (BHI, Millipore, 70138)and allowed to grow overnight. Biological triplicates as well as technical duplicates were made to allow for statistical analysis.
Staphylococcus aureus NCIMB 13062 was used. For comparative purposes, Escherichia coli NCIMB 12805 was also included as well. Additional data was run for Salmonella enterica spp. enterica NCIMB 13284 but data is not included here. It is available upon reasonable request.
10 μL of the overnight S. aureus culture was inoculated into each well of a 96-well microtiter plate (not including the negative media controls), and subjugated to different nutrient concentrations at (19%, 15%, 10%, 5%, 2.5%, and 0% w/w). This was done for 3 different media, namely Tryptic soy broth (TSB, Neogen, NCM0019A), potato dextrose broth (PDB, Neogen, NCM0157A) and brain heart infusion broth (BHI, Millipore, 53286.
Each 96-well plate was run either in the RID or the plate reader (Epoch 2, SN 16121910).Measurements were done every 1 hour for 48 hours.
In the RID the overall score of turbidity was measured every hour for 48 hours total. Parameter exported was top-light value score (HSV). Alternatively, bottom light could have been used, but upon data inspection, this seemed to have lower sensitivity and was therefore discarded. In the plate reader, the optical density, OD600 (𝜆=600) was measured. The assay was run at 25°C (RT),32°C, 37°C and 40°C.
Results
Samples were normalized so that at T=0, all first measurement values were converted to 1.For ease of visualization, only data from the 40°C was shown here. All other data is available upon reasonable request.
The following curves were obtained for the assessment of growth of S. aureus NCIMB 13062.
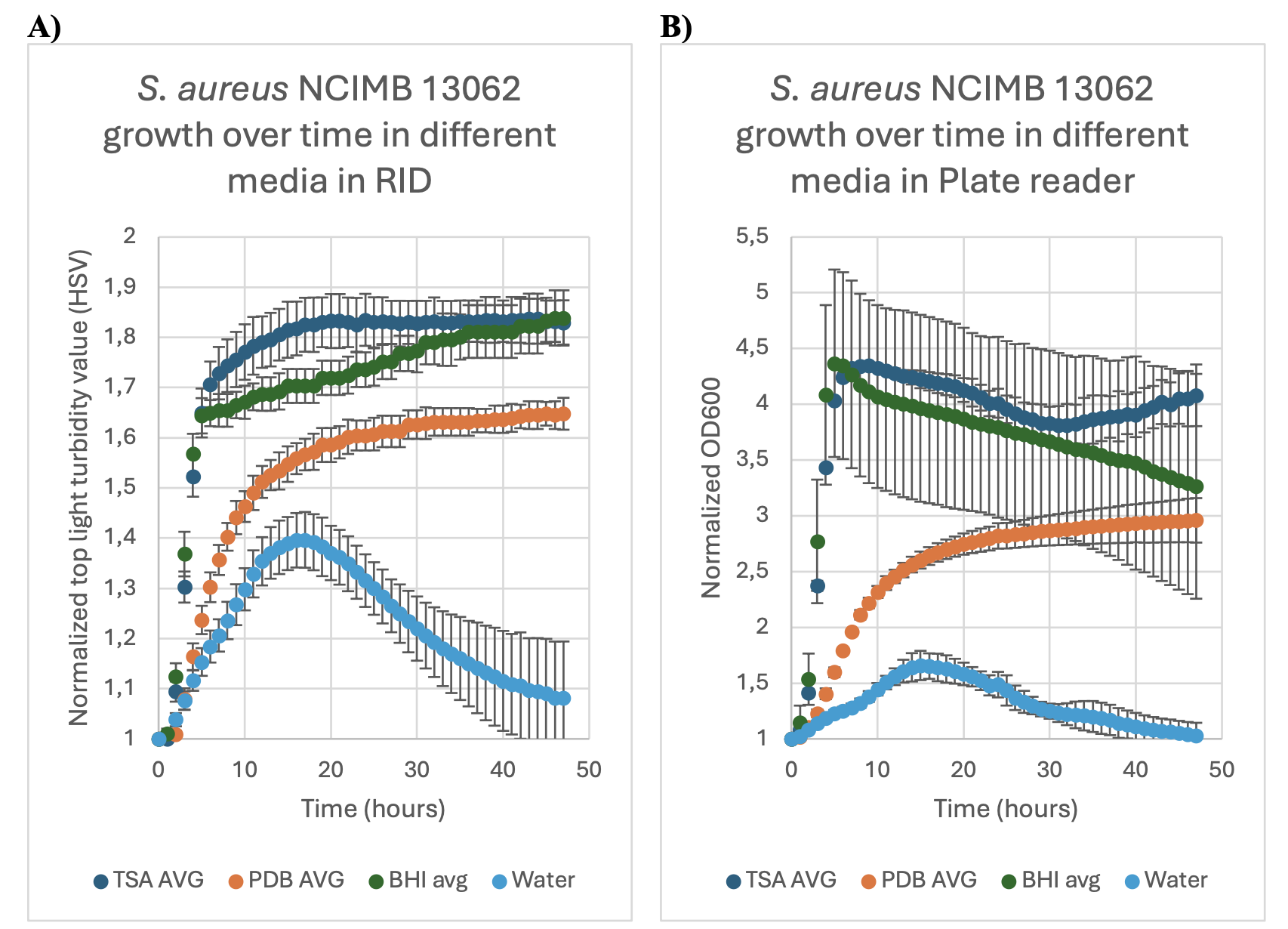
FIGURE 1: The growth (normalized turbidity and OD600 measurements) over time for S.aureus NCIMB 13062 in three different media; Tryptic soy broth (TSA AVG), Potato dextrose broth (PDB AVG) and brain-heart infusion broth (BHI AVG) and water, over the time span of48 hours. Error bars indicate the standard deviation of the samples as calculated from the technical duplicates and biological triplicates. Experiment was conducted at 40 °C. 1A: in theReshape imaging device (RID). 1B: in plate reader (Epoch 2).
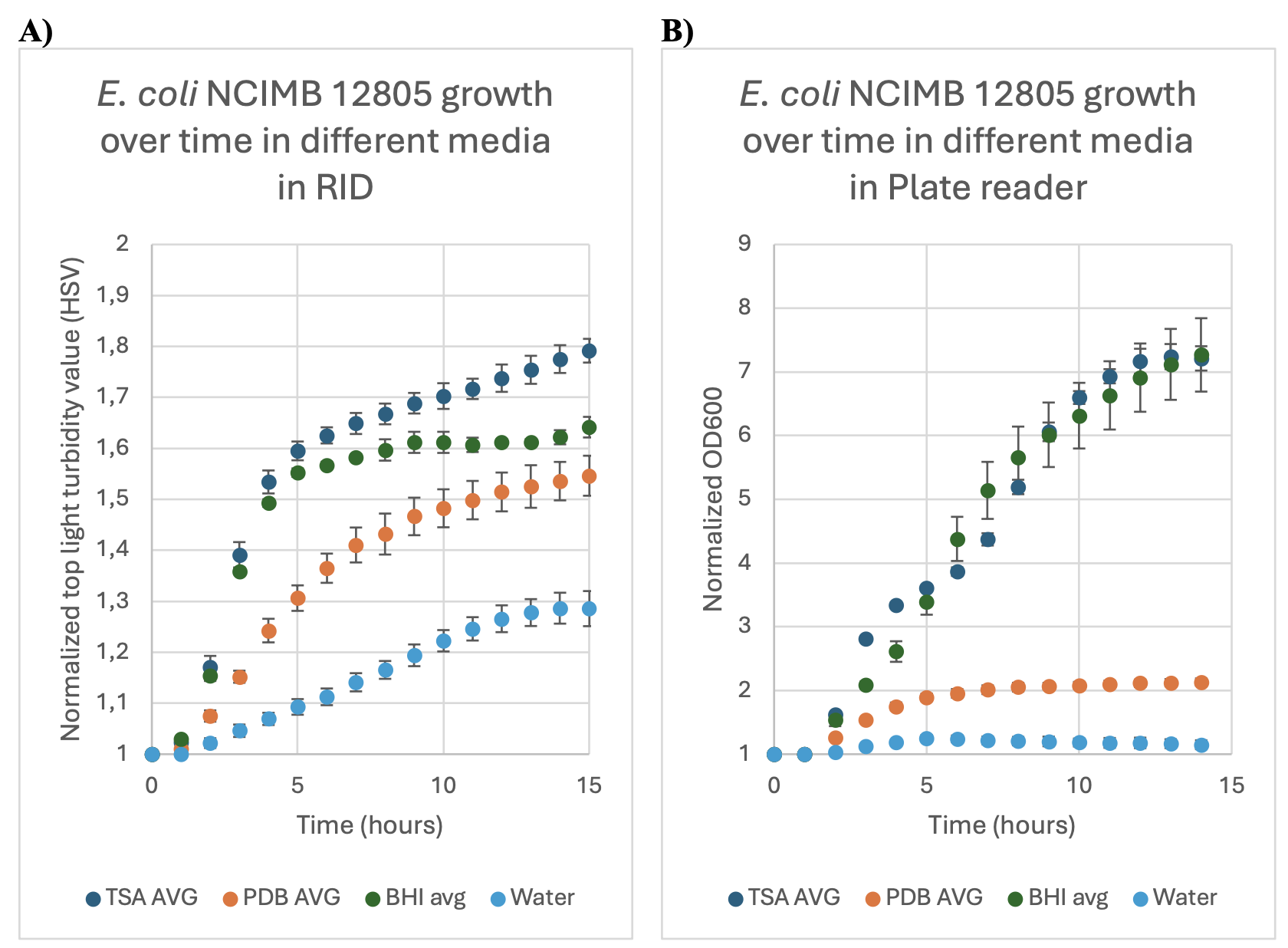
FIGURE 2: The normalized growth (as expressed through turbidity and OD600measurements) over time of E. coli NCIMB 12805 in three different media, Tryptic say broth(TSA AVG), potato dextrose broth (PDB AVG) and brain heart infusion broth (BHI), andwater, over the time span of 14 hours. Error bars indicate the standard deviation of the samples as calculated from the technical duplicates and biological triplicates. Experiment was conducted at 40 °C. 2A: in the RID, 2B: in plate reader (Epoch 2).
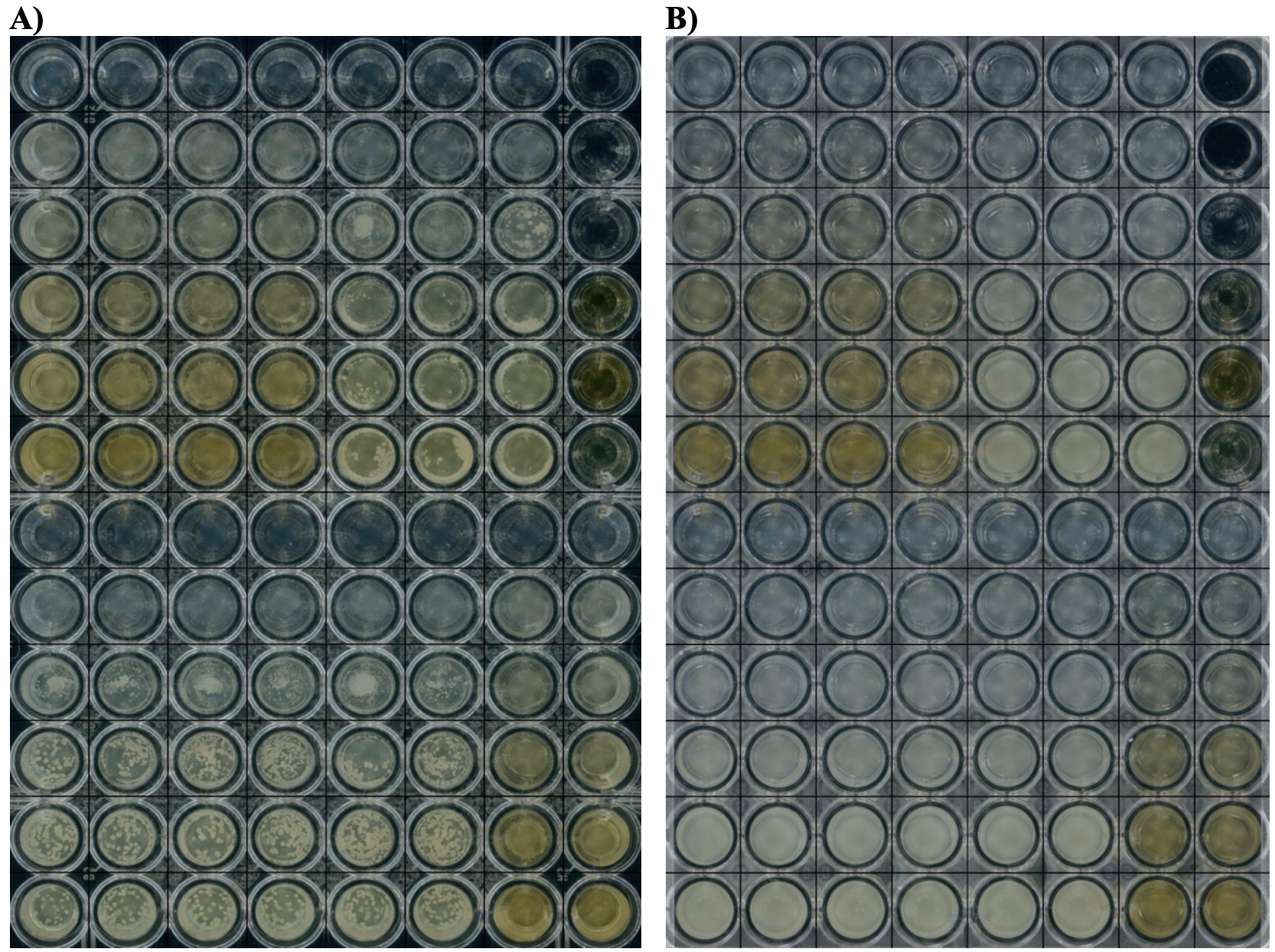
FIGURE 3A: Visual images after 48 hours as seen in the RID of S. aureus NCIMB 13062 in a96-well plate used for growth assay using top light setting. S. aureus grows in very visible aggregates. FIGURE 3B: Visual images after 48 hours as seen in the RID of E. coli NCIMB12805 in a 96-well plate used for growth assay.
Discussion: Comparative Analysis of RID and Plate Reader
Initially looking at the data generated from the RID and the plate reader and trying to compare the two results in the following points (1 through 7), and is summarized in point 8, the table at the end.
1. Performance on Low-Value Measurements
One notable area where the plate reader excels is in low-value measurements, such as those close to the baseline (e.g., water or very dilute cultures). In these cases, the plate reader consistently shows lower standard deviations (stdev) between replicates, suggesting it offers more reliable signal detection at the bottom end of the dynamic range. This may be due to its finely tuned optical path and background correction algorithms optimized for low turbidity readings.
In contrast, the RID (Reshape Imaging Device) tends to show higher variance at these low values, possibly due to noise amplification or limited sensitivity near zero. For precision atextremely low cell densities, this can present a limitation. This generally indicates that the RID has less resolution as compared to the plate reader.
2. Superior Consistency in Higher Ranges: RID Advantage
Outside the low-signal regime, however, the RID demonstrates significantly lower standard deviation in turbidity measurements. Across the range of cell concentrations tested (exceptwater), RID consistently outperforms the plate reader in terms of reproducibility and noise suppression. This suggests a strong case for RID in mid-to-high turbidity applications, where robust data is crucial, and variability can skew biological interpretations.
3. The Case of S. aureus vs. E. coli
A notable biological factor is that S. aureus aggregates wildly, creating inconsistencies in optical-based systems due to uneven light scattering. This can artificially inflate or deflate readings depending on clump orientation and size. As a comparison, E. coli does not co-aggregate to the same extent, offering a more stable reference curve.
Data analysis comparing the two organisms shows that RID remains more consistent even in the presence of aggregation, potentially due to how it integrates imaging data over a larger area or time window. Drawing this comparison highlights RID’s resilience to biological noise—a major strength.
4. Curve Trends: Consistency Across Devices
Importantly, despite absolute value differences, the overall shape and trajectory of the growth/turbidity curves remain consistent between RID and plate reader. This means that biologically relevant trends (growth phases, inflection points) are preserved in both systems, reinforcing RID’s potential as a high-throughput alternative.
5. Value in Bulk Characterization
For bulk assays—where minute differences are less critical and trends over large sample sets matter—RID offers massive advantages. The ability to process up to 10 plates simultaneously drastically reduces run time and enables higher experimental throughput, which is a game changer for screening pipelines or production QC environments.
6. Sensitivity Considerations: Applicability to MIC Assays?
A potential limitation of RID is sensitivity in detecting small concentration changes, such as in Minimum Inhibitory Concentration (MIC) assays. Since these depend on resolving subtle shifts near the threshold of inhibition, the current sensitivity of RID might not be adequate unlesscarefully calibrated. This is especially critical when precise quantification determines biological outcomes. In the future this will require further validation concerning the sensitivity of the measurements.
7. Media Color Effects: A Confounding Variable
One theoretical concern for RID (and to a lesser extent plate readers) is color interference from growth media. Colored media could alter how light is absorbed or scattered, leading to false positives or skewed turbidity signals. While plate readers can sometimes correct for this via dual-wavelength or reference filters, further testing is needed to confirm whether RID can handle such variability or if certain media types should be avoided.
8. Comparative Overview and Drawbacks
Both systems have their trade-offs. While plate readers are better for low-end sensitivity, RID clearly shines in consistency, scalability, and potential robustness to sample aggregation.
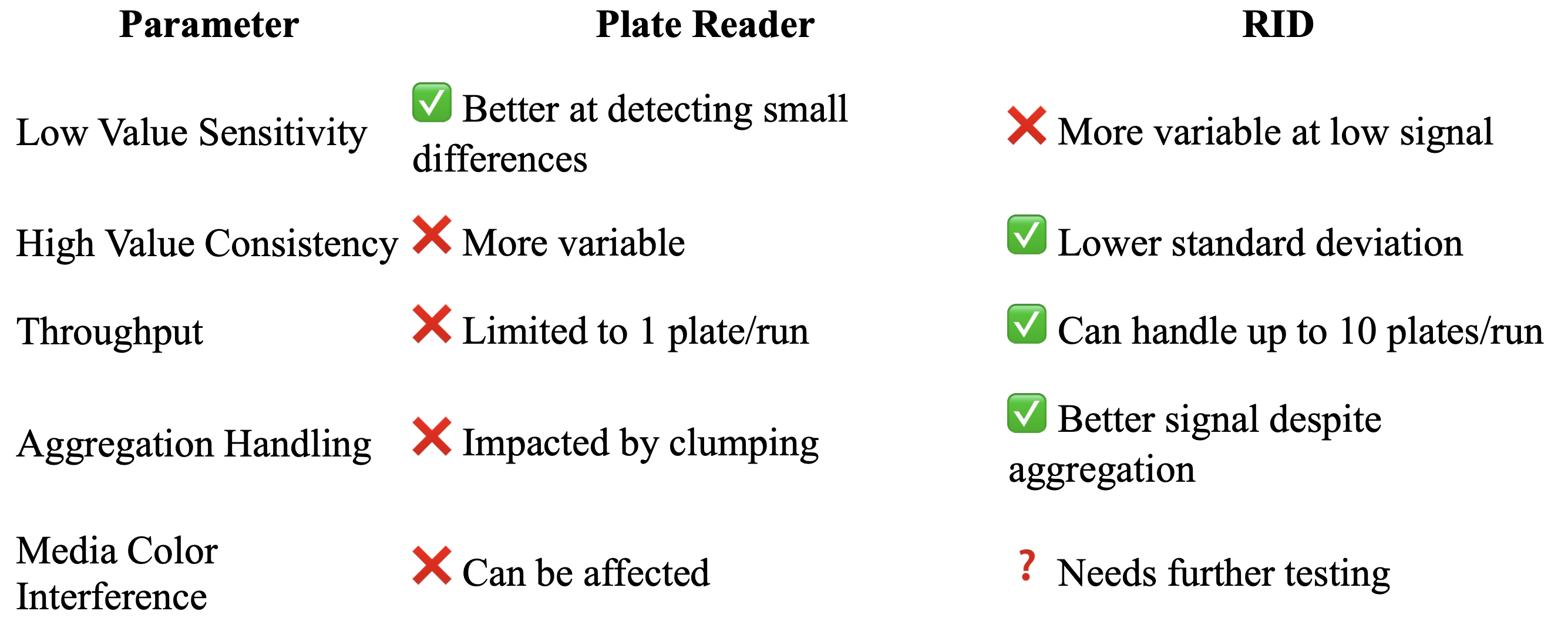
Both systems have their trade-offs. While plate readers are better for low-end sensitivity, RID clearly shines in consistency, scalability, and potential robustness to sample aggregation.
Conclusion
In summary, while plate readers maintain their edge in low-end sensitivity and some standardization, the RID presents a compelling alternative, particularly for bulk and mid-range turbidity assays. It combines lower variance, higher throughput, and potential resilience to sample heterogeneity. With further validation and calibration—especially for sensitive applications like MIC determination—RID could revolutionize cell-based screening workflows.
With this kept in mind, the RID offers a viable alternative to the plate reader, and as it is a cheaper alternative to the plate reader (potentially also with the pay-os-you-go scenario) it will in the future lower the bar for start-ups trying to get proof of concept on their ideas.
Future perspectives
Lower economic input needed to proof of concept will enable both smaller and bigger companies to test ideas in a. high-throughput manner, and more easily decide which concepts show potential. Additionally, the visual validation of the plate in a “hands-off” manner allows for easier troubleshooting when setting up assays. There is a need for further validation though, which is on-going at Reshape Biotech.
Personal care & Cosmetics
Automate testing and analysis of cosmetics and personal care products to ensure product quality, safety, and efficacy.
Read more
Industrials
Enable automated high throughput microbial discovery or do library and enzyme screens with much higher resolution.
Read more
Food & Ingredients
Supercharge your functional ingredient characterization, stability testing, acidification, and other assays.
Read more
Agriculture
Accelerate agricultural R&D with automated Imaging and AI-powered image analysis
Read more

